Science
How Forensic Science Helps Solve Murder Mysteries
During the investigation, the team focused on proteins of non-human origin.

In 2014, a Canadian coroner came across an odd murder case: The unfortunate homicide victim had uniform black and blue marks all over her body. However, there was no indication of blunt force trauma, and the skin showed no obvious wounds. The Canadian police decided to contact a university professor who was an expert in protein science. What happened next created history in the world of forensic proteomics.
Forensic proteomics unleashed
The professor’s research laboratory analyzed the blood sample obtained from the victim’s body using a variety of analytical techniques. After eliminating some of the most abundant proteins present in human blood, the research team focused on proteins of non-human origin. During their extensive search, they came across a likely candidate: an enzyme called phospholipase A2. Phospholipase A2 or “PLA2” is a protein that possesses enzymatic activity. It hydrolyzes the ester bonds present in membrane phospholipids, thus producing a free fatty acid such as arachidonic acid and a lysophospholipid. As we all know, arachidonic acid triggers a cascade of biochemical processes that cause tissue damage in multiple body organs.
Equipped with this new evidence, the Canadian police obtained a warrant to thoroughly searched the residence of the prime suspect. Upon searching, they found a variety of exotic snakes. The court issued a sentence based on all of this evidence. This landmark case opened new avenues for the field of forensic proteomics. The related study got published in the journal Forensic Science International.
What is forensic genealogy?
Nabbing a deadly murder suspect or a serial killer is definitely not an easy task. If the suspect has no previous criminal history, it means that law enforcement does not have his or her DNA profile available for cross-checking against crime scene DNA samples. This complicates the entire investigative process. However, thanks to a new forensic technique, cops are now able to zero in on potential murder suspects much easily than ever before. The technique, known by the name “forensic genealogy,” involves the following steps:
- Crime scene investigators collect DNA samples from the crime scene.
- The DNA samples are then sequenced.
- The generated DNA profiles are then uploaded to a public DNA database that people generally use to facilitate genealogy studies.
- Depending on the extent of similarity, law enforcement can interrogate the identified relatives of the potential suspect.
- Smart detectives then pool together all the data and try their best to arrest the deadly suspect.
You will be surprised to know that many deadly suspects who had evaded detection were arrested purely based on forensic-genealogy-based analyses. However, this technique has two major limitations:
- People are increasingly asking genealogy websites to shield their DNA profiles from law enforcement (see the second reference at the end of this article).
- Most people rarely have their own DNA profiles ready with them.
Let’s hope that more and more people voluntarily submit their DNA profiles to open-source DNA registries in the near future.
Please share your thoughts in the comments section of the embedded videos or on this page. By the way, what is your opinion on the D. B. Cooper mystery?
Key takeaways
- Forensic proteomics and forensic genealogy are promising fields with bright prospects.
- Why kill someone? Live and let live.
Main references
Journal: Forensic Science International | Title: The application of forensic proteomics to identify an unknown snake venom in a deceased toddler | DOI: 10.1016/j.forsciint.2021.110820
Journal: Science | Title: Regulating forensic genetic genealogy | DOI: 10.1126/science.abj5724
Science
CRISPR-Like System Discovered in Animals
The newly discovered/engineered genome editor has clinical implications

“CRISPR” (Clustered Regularly Interspaced Short Palindromic Repeats) and “Cas” (a CRISPR-associated “endonuclease,” or an enzyme that cuts the phosphodiester bonds present between the nucleotides of nucleic acids) became familiar phrases during the last decade. At present, the CRISPR-Cas system is being used extensively in research laboratories across the globe to perform custom targeted DNA editing. The CRISPR “defense system” was originally discovered in genomes of bacteria and archaea. Its primary function is to serve as a “memory reservoir” of past infections.
So, what’s new? Well, MIT and Harvard researchers have recently discovered CRISPR-like systems even in animals.
Why do organisms need CRISPR?
When cops arrest a criminal, they fingerprint him/her. Similarly, when a virus/plasmid infects a bacterium, the latter captures a small segment of the invader’s nucleic acid sequence. The bacterium then uses this tiny segment to create a permanent “criminal record” in the CRISPR system. This record is then used to protect the bacterium from future attacks. If the intruder strikes again, the bacterium activates its CRISPR “defense system.” Upon activation, the CRISPR system cleaves the nucleic acid sequence of the intruder. It does this by using the molecular scissors of the Cas enzyme. That’s exactly how the “smart” bacterium protects itself from “serial” offenders.
Although such systems were known to exist in lower organisms such as bacteria and archaea, their presence has now been detected in higher organisms. The associated report has been published in the high-impact journal Nature.
What is “Fanzor” and what will it do?
In the recently published Nature report, the research team from MIT and Harvard shows how a protein called “Fanzor” can use RNA as a guide for modifying DNA with extreme efficiency. The report also discusses how Fanzors can be reprogrammed to edit the genome of human cells. According to the research team, this landmark discovery paves the way for therapeutic applications resulting from highly efficient human genome editing.
“Fanzor,” the first RNA-guided DNA-cutting enzyme found in eukaryotes, could one day be harnessed to edit DNA more precisely than CRISPR/Cas systems.
Did they conduct any “proof-of-concept” experiments?
Yes, the researchers demonstrated the use of Fanzor as a genome editing tool. They did this by generating modifications at targeted genome sites within human cells. Moreover, they systematically engineered Fanzor by introducing key mutations into it after discovering that the Fanzor system was less efficient at cleaving DNA than conventional CRISPR-Cas systems. These mutations increased the activity of Fanzor 10-fold. Long story short, we now have a highly efficient “genome editor” in our possession. Fingers crossed!
Reference: Nature | DOI: 10.1038/s41586-023-06356-2
By the way, did you know that space scientists have discovered carbon in the Orion Nebula?
SciTechVault.com reviews, simplifies, and communicates research that matters. We diligently go through published (high-quality) scientific research from across disciplines, understand the underlying concepts, and authoritatively communicate the same to our target audience in a unique, simplified, and engaging manner. We also create scientific content for our YouTube channel. Did you get a chance to check out some of our recent scientific communications?
Science
Genetically Altered Fungi Love Eating Plastic
Genetically modified Aspergillus fungi eat plastic and convert it into pharmacologically active compounds

Amelia was a nature lover. She used to enjoy hiking and wildlife photography. Amelia used to take special efforts to minimize her individual carbon footprint. She used to carry her own grocery bags to the nearby grocery store. Quite recently, Amelia came across an interesting study published in the reputed journal Angewandte Chemie. She felt delighted after reading the peer-reviewed article.
Can fungi eat plastic?
Polyethylene is notorious for a reason: It is not easily biodegradable. Researchers from the University of Kansas recently demonstrated that genetically engineered strains of Aspergillus nidulans are able to consume polyethylene-based plastic and convert it into useful metabolites. These metabolites exhibit properties that are important from the pharmacological point of view, according to the study. The pharmacologically significant compounds include asperbenzaldehyde, citreoviridin, and mutilin. Chemical derivatives of mutilin, for instance, are used as antimicrobial agents.
Quite interestingly, a previous study has shown that certain bacteria can also extend similar help. The study focuses on the biodegradability of polyethylene by bacteria derived from the guts of plastic-eating waxworms.
Why does A. Nidulans produce useful compounds?
According to the researchers, the genetically modified A. nidulans produces useful chemical compounds with pharmacological significance primarily to protect itself from other organisms or to compete with them. For instance, it is an established fact that the Penicillium mold produces “Penicillins” (a group of antibiotics that harbor the famous “β-lactam ring”) to defend itself against microbes. Did you know that β-lactam-based antibiotics kill bacteria by inhibiting the biosynthesis of bacterial cell walls? Well, you know it now.
This is an extremely important finding, because it can effectively address the issue of environmental pollution caused by discarded plastic items and that’s exactly why the researchers were able to publish it in a high-profile journal. The journal Angewandte Chemie has a Jan. 2023 h-index of 579!
Key takeaways
- We now have in our arsenal, a novel method for converting polyethylene-based plastic into chemical compounds that are of great significance to the pharmaceutical industry.
- Nevertheless, we must all draw inspiration from Amelia and minimize our use of plastic.
Main reference
Journal: Angewandte Chemie | Title: Conversion of Polyethylenes into Fungal Secondary Metabolites | DOI: 10.1002/anie.202214609
By the way, did you know that freshwater fish caught in the U.S. produce the notorious “forever chemicals”?
Science
Solar-Powered Worm with an Extended Lifespan
The published study says that the genetically altered worm generates chemical energy (ATP) in the presence of sunlight
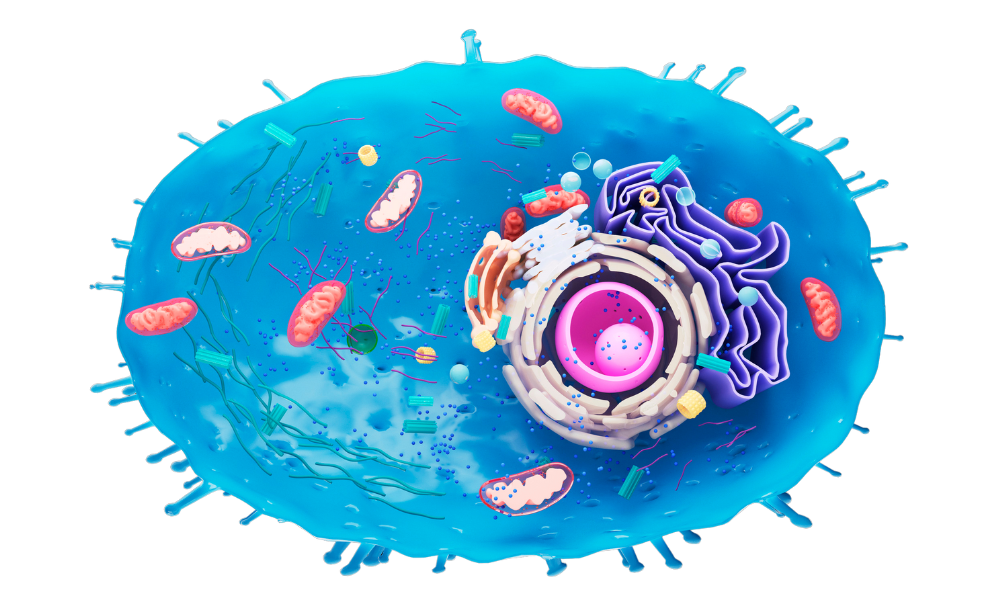
Folks at SciTech Vault love science fiction movies. Also, the title of this article sounds too good to be true; almost like a sci-fi movie from Hollywood. However, you will be surprised to know that we haven’t exaggerated anywhere at all. A landmark study published in Nature Aging indeed shows that genetically modified Caenorhabditis elegans (C. elegans) that harnesses solar energy lives 30 to 40% longer than its regular (non-solar) counterpart.
How did that happen?
Well, researchers from the United States did something unusual: They genetically modified the C. elegans mitochondria by making them incorporate a light-activated proton pump. The light-activated proton pump was sourced from a fungus. As we all know, mitochondria are specialized cellular organelles that generate (chemical) energy in the form of adenosine triphosphate or “ATP.” Thanks to the newly added proton pump, the genetically altered C. elegans started generating additional energy by harnessing sunlight. Moreover, the integration of this “solar” pump boosted its lifespan by up to 40% (please see the illustration below).

Underlying mechanism
The genetically incorporated proton pump moves charged particles across the mitochondrial membrane in the presence of sunlight. In other words, it “charges” the mitochondria by harnessing solar energy. As a result, the genetically altered nematode witnesses an extended lifespan presumably because its mitochondria receive “supplemental” or extra support from the solar-powered proton pump. In other words, the organism receives an extra boost of energy from the genetically incorporated proton pump, and this boost (presumably) makes it live longer than usual.
On a related note, C. elegans serves as a good model organism because of various reasons. For instance, the organism shares a relatively high genetic homology with humans. It can be economically cultured in large quantities. Moreover, the relatively short lifespan of C. elegans drastically facilitates the rapid screening of anti-aging compounds.
By the way, did you know that scientists recently discovered cyclic peptides with chemotherapeutic potential?
Key takeaways
- We now have an animal model showing that it is entirely possible to generate energy (and extend lifespans) by utilizing solar energy.
- Thanks to the numerous advances in science, the life expectancy of human beings is also increasing steadily. No, we won’t be coming across solar-powered humans anytime soon. However, it is important to note that people (who make good lifestyle choices) are able to live relatively long lives these days. Cheers (but without any alcohol, ok?)!
Main reference
Journal: Nature Aging | Title: Optogenetic rejuvenation of mitochondrial membrane potential extends C. elegans lifespan | DOI: 10.1038/s43587-022-00340-7
Science
Cyclic Peptides Target Cancer Cells
Researchers from Israel and Japan discover novel cyclic peptides with chemotherapeutic potential
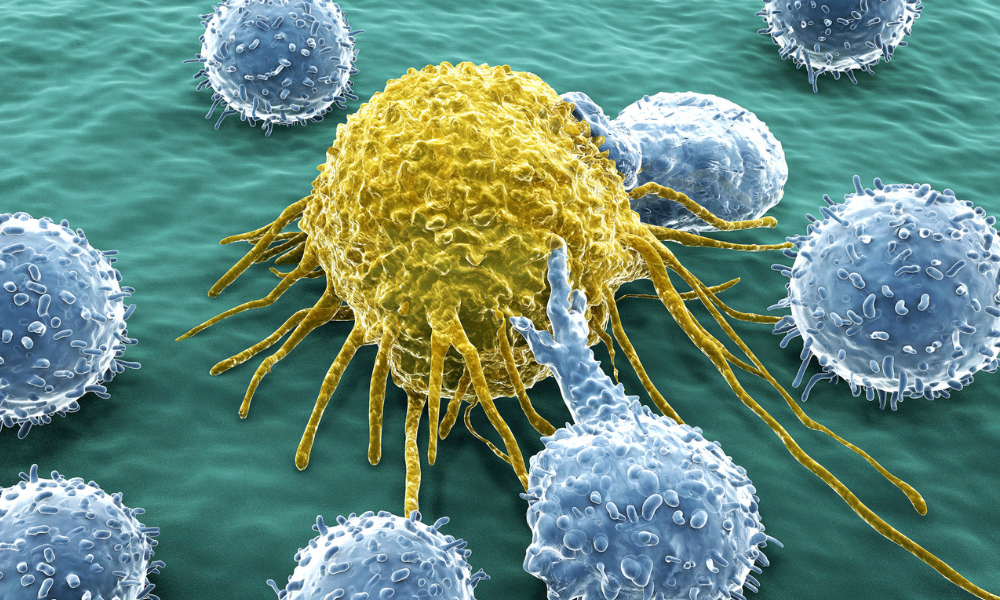
Humanity’s fight against cancer continues. Funding agencies have funded research grants worth billions of dollars in an attempt to find high-quality treatments for cancer. Of course, scientists from around the world have already made extremely remarkable and noteworthy contributions to the area of cancer prevention and chemotherapy. An international team of researchers from Israel and Japan has recently published a landmark study in Nature Communications. The study focuses on the role of novel “cyclic peptides” as attractive candidates for cancer treatment.
Cyclic peptides and their mechanism of action
Unlike proteins—large biomolecules comprising amino acids that are connected via peptide bonds—cyclic peptides consist of only a few amino acids that form a circular structure. Famous examples include bacitracin (an antibiotic) and cyclosporin (an immunosuppressant).

The researchers from Japan and Israel discovered cyclic peptides that bind to the protein “ubiquitin;” a protein that carries out important functions such as protein degradation/recycling and DNA repair, without which it is difficult for cells to survive.
Long story short, the researchers used the newly discovered cyclic peptides to target ubiquitin inside cancerous cells. When the cyclic peptides bind ubiquitin chains at a particular location, they inhibit the process of protein degradation/recycling. When this happens inside a cancerous cell, it ends up killing the rogue cell. This epic discovery resulted into a Nature Chemistry paper in the year 2019.
Quite recently, instead of targeting the process of protein degradation/recycling, the research team targeted the process of DNA repair inside cancerous cells and used it to demonstrate the chemotherapeutic potential of cyclic peptides.
How does ubiquitin facilitate protein recycling?
“Ubiquitin” is a protein known for its ability to “fire” unwanted proteins from their job. By attaching itself to unwanted proteins, ubiquitin directs them to a famous address—a protein complex known as the “proteasome.” Once inside this complex, the “fired” proteins carrying the ubiquitin tag get degraded and recycled into new proteins. Nature has devised many such awe-inspiring mechanisms of action in a variety of organisms.
Key takeaways
- We now have a promising approach for chemotherapeutic intervention. Further research is warranted.
- Physicians prescribe medicines but rarely know their detailed mechanisms of action (or how they are synthesized). Let’s take this opportunity to give a standing ovation to biochemists and organic chemists; scientists who (actually) discover mechanisms and design therapeutic drugs.
On a completely different note, women possess more empathy and can read minds more efficiently than men.
Main reference
Journal: Nature Communications | Title: Selective macrocyclic peptide modulators of Lys63-linked ubiquitin chains disrupt DNA damage repair | DOI: 10.1038/s41467-022-33808-6















































